Jordan Rozum
I am a postdoctoral computational biologist at Pacific Northwest National Lab. This page primarily describes my work prior to joining PNNL.
Prior to joingin PNNL, I was a postdoctoral research associate studying complex systems in the Complex Adaptive Systems & Computational Intelligence (CASCI) laboratory at Binghamton University. I have a math and physics background, and a research focus on applications to systems biology.
Research Interests
Current work: systems biology modeling, machine learning applications to protein structure, complexity science, network dynamics
Previous work: social networks in healthcare, galaxy modeling, airglow physics, differential geometry
I have a math and physics background, and my current work focuses on applications of data science and dynamics to systems biology. Much of my work has been about how feedback loops in biological networks give rise to complex behaviors. I’ve applied these ideas in various contexts, including cell cycle regulation, pattern formation during embriogenesis, and immune cell signaling. I enjoy working with Boolean networks, which are discrete dynamical models popular in systems biology.
I am the lead developer of the Python libraries pystablemotifs and cubewalkers, which together allow for fast attractor detection, control, and GPU-accelerated simulation in Boolean networks. I apply these methods to study the target control problem in experimentally-derived models and order-to-chaos phase transitions in random networks.
Recently, I have also been developing methods to sparsify multi-layer networks while taking into account multiple data sources. I apply these methods to various problems in computational social science, health, and biomedical complexity.
Highlights: Characterizing robust behaviors in cells
“Models of cell processes are far from the edge of chaos”
KH Park, FX Costa, LM Rocha, R Albert, JC Rozum
PRX Life 1, 023009, 2023
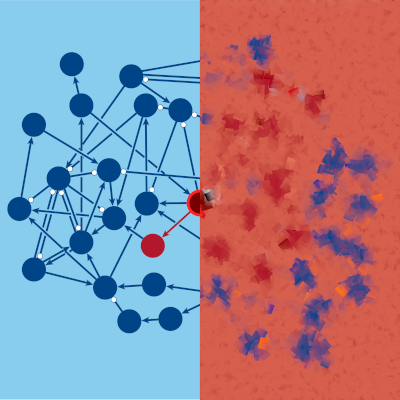
Powerful new simulations of cell processes models using our Python library cubewalkers have revealed more robustness than previously proposed, challenging the prevailing “edge of chaos” theory (simulation code and results available here). The edge of chaos theory is based on the idea that life needs to adapt to its environment without being hypersensitive to it. If cells behave chaotically, then the tiniest errors can snowball into catastrophic–and fatal–failure. On the other hand, if they behave with rigid, crystalline order, then they are slow to adapt to changes in their environment. The edge of chaos is the critical point that separates these regimes and is thought to be the “sweet spot” where living systems operate. That was not the case here. In one of the largest databases of experimentally-supported cell process models, cells were able to recover from perturbations with much more resiliency than the edge of chaos hypothesis predicts. This suggests that, at least in the processes studied, life is willing to sacrifice some adaptive flexibility in favor of day-to-day robustness, challenging the notion that cells live life on the edge of chaos.
See the relevant paper here
“Identifying (un) controllable dynamical behavior in complex networks”
JC Rozum and R Albert
PLoS Computational Biology 14 (12), 2018
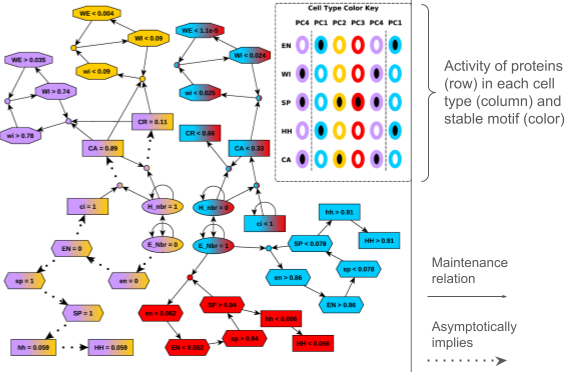
We developed a formalism that identifies positive feedback loops in ODE models that can “lock in”, causing a system to commit to a restricted range of possible long-term behaviors. We used this formalism to analyze the biomolecular decisions that lead to the formation of embryonic segments in the fly (Drosophila melanogaster) and to uncover a self-reinforcing feedback loop in the biomolecular circuitry that governs the response of T-cells to external signals. In both cases, we showed how these methods identify manipulations of the system that can disrupt observed phenotypes and manipulations that cannot. Apart from their utility in basic science research, observations of this nature have important potential applications in the development of pharmaceuticals because they can narrow the (often very expensive) search for potential drug targets.
See the relevant paper here.
Highlights: Understanding the biomolecular decisions that cells make
“Parity and time-reversal elucidate both decision-making in empirical models and attractor scaling in critical Boolean networks”
JC Rozum, JGT Zañudo, X Gan, D Deritei and R Albert
Science Advances 7 (29), eabf8124, 2021
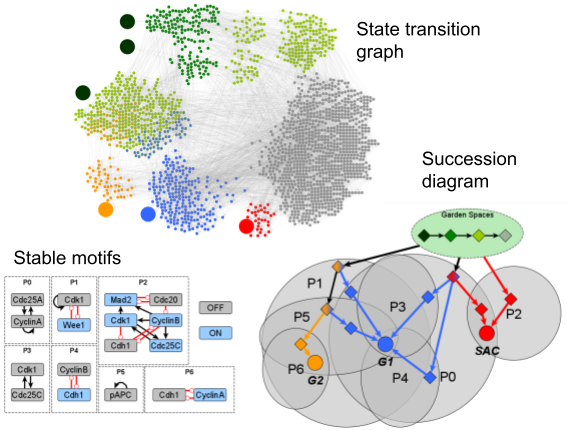
We developed the open-source Python library pystablemotifs, which provides one of the world’s fastest algorithms for attractor identification and control in Boolean networks. This library uses parity and time reversal provide new insights into the key regulatory circuits that underpin the biochemical “decisions” that cells make. We used this tool to identify the attractor repertoires of networks 80 times larger than ever before considered and thereby resolved a 50-year-old open problem about how stochasticity affects a cell’s diversity of phenotypes. This was the first time that stochastic genome-scale biomolecular models were analyzed in such detail. The results of our analysis showed that the behaviors of traditional random models of gene regulation are highly sensitive to stochasticity, suggesting biological selection pressures that are not accounted for in the canonical model.
See the relevant paper here, and a popular press summary here.
Research Experience
Postdoctoral Research Associate 2022-Present, Binghamton University (SUNY) Dept. of Systems Science and Industrial Engineering
I am a current member of the Complex Adaptive Systems & Computational Intelligence (CASCI) laboratory lead by Professor Luis Rocha. I work on developing new tools and algorithms for analyzing complex networks in a variety of domains, including social media, biomedical data, and systems biology. I apply these tools to gain new insights into robustness and redundancy in biomedical systems from the cellular scale to the social scale.
Graduate Research Assistant 2016–2022, Pennsylvania State University Dept. of Physics
Developing new network control approaches that generalize methods for discrete dynamics to continuous ODE network models in collaboration with Professor Réka Albert. Apply network control theory to biological network models to better understand decision mechanisms in cellular systems, identify potential drug targets, and predict results of biological experiments.
Graduate Research Assistant 2014-2016, Utah State University Dept. of Mathematics
Classification of homogeneous space-times according to their symmetries as part of Master’s Thesis advised by Professor Ian Anderson and Professor Charles Torre. Devised a new coordinate-free classification scheme for Lie algebras of a certain type relevant to homogeneous solutions to the Einstein equations and implemented the classification using the DifferentialGeometry package of the computer algebra system Maple.
Research Programmer, 2010-2014, Utah State University Dept. of Electrical Engineering
Data validation for the SABER (Sounding of the Atmosphere using Broadband Emission Radiometry) satellite instrument in collaboration with Professor Gene Ware and Professor Doran Baker. Analyzed SABER data, and maintained the USU copy of the database. Focused on perceived bifurcation of hydroxyl airglow layer in the mesosphere.
Undergraduate Research Fellow, 2010-2014, Utah State University Dept. of Physics
Examining bias in galaxy catalogs due to galaxy geometry in collaboration with Professor Shane Larson as part of USU’s Undergraduate Research Fellowship. Wrote computer simulations of galaxy surface brightness. Performed statistical analyses of galaxy catalogs to search for possible indications of observation bias.
Education
Ph.D. in Physics from Pennsylvania State University, 2022
- Concentration: Network biophysics and dynamical systems
- Dissertation: Attractor identification and control of Boolean and ODE network models in Systems Biology
- Advisor: Professor Réka Albert
Master’s in Mathematics from Utah State University, 2016
- Concentration: Mathematical physics and differential geometry
- Thesis: Classification of Five-dimensional Lie Algebras with One-dimensional Subalgebras Acting as Subalgebras of the Lorentz Algebra
- Advisors: Professor Ian Anderson and Professor Charles Torre
Bachelor’s in Physics and Mathematics from Utah State University, 2014
- Magna Cum Laude
- Capstone project: Analyzing solutions to the Einstein equations using the Maple package DifferentialGeometry
Honors, Awards, and Society Memberships
- PSU ICDS Featured Researcher (link), 2021
- PSU Peter Eklund Memorial Lectureship Award Honorable Mention, 2019-2020
- PSU David Duncan Graduate Fellowship ($1650), 2019
- PSU Department of Physics David H. Rank Memorial Award ($1200), 2017
- PSU College of Science Roberts Scholarship Award ($2000), 2016
- USU Magna Cum Laude, 2014
- USU Physics Department Outstanding Senior, 2014
- Barry M. Goldwater Scholar ($7500), 2013
- Barry M. Goldwater Scholarship Honorable Mention, 2012
- USU A-Pin Award for Consecutive 4.0 Semesters, 2010-2014
- USU Undergraduate Research Fellow ($4000), 2010-2014
- USU Presidential Scholar (tuition & fees, approx. $40,000), 2010-2014
- Sigma Pi Sigma honor society member
- NetSci member
Teaching Experience
Teaching Assistant, Pennsylvania State University Department of Mathematics 2016-2017
Course: Introductory Classical Mechanics
Course Instructor, Utah State University Department of Mathematics 2015
Course: Calculus II
Teaching Assistant, Utah State University Department of Mathematics 2015
Courses: College Algebra
Teaching Assistant, Utah State University Department of Physics 2013-2014
Courses: Introductory Physics I and II
Undergraduate Teaching Fellow, Utah State University 2011-2013
Courses: Advanced Classical Mechanics, Advanced Electromagnetism I and II
Reviewer
Bioinformatics
Biophysical Journal
BMC Bioinformatics
Journal of Complex Networks
Journal of Mathematical Biology
Mathematics in Science and Industry
Mathematical Biosciences and Engineering
Nature Communications
npj Systems Biology and Applications
Physica D: Nonlinear Phenomena
PLOS Computational Biology
Scientific Reports
Skills
Programming expertise in Python, additional completed projects using C++, CUDA, MATLAB/Octave, R, Rust, Sage, others
Lead developer of pystablemotifs (Python) and cubewalkers (Python+CUDA) libraries
Expertise in network science and discrete dynamical modeling
Strong background in mathematics and algorithm development
Experience with high-performance computing and managing large datasets
Publications
in preparation
AM Marcus, JC Rozum, H Sizek, LM Rocha. CANA 1.0 and schematodes: efficient quantification of symmetry in Boolean automata (working title). In preparation.
Z Guo, J Felag, RB Correia, JC Rozum, LM Rocha. Selection of relevant patient cohorts from social media using the metric backbone of knowledge networks. In preparation.
JC Rozum and LM Rocha. The ultrametric backbone is the union of all minimum spanning forests. In preparation.
submitted
KH Park, JC Rozum, R Albert. From years to hours: accelerating model refinement. Submitted.
JC Rozum, C Campbell, E Newby, FSF Nasrollahi, R Albert. Boolean Networks as Predictive Models of Emergent Biological Behaviors. Accepted, Cambridge Elements (2023)
published
KH Park, FX Costa, LM Rocha, R Albert, JC Rozum. Models of cell processes are far from the edge of chaos. Accepted, PRX Life 1, 023009 (2023)
R Li, JC Rozum, MM Quail, MN Qasim, SS Sindi, CJ Nobile, R Albert, AD Hernday. Inferring gene regulatory networks using transcriptional profiles as dynamical attractors. PLoS Computational Biology 19 (8), e1010991 (2023)
FX Costa, JC Rozum, AM Marcus, LM Rocha. Effective Connectivity and Bias Entropy Improve Prediction of Dynamical Regime in Automata Networks. Entropy 25 (2), 374, (2023)
JC Rozum and R Albert. Leveraging network structure in nonlinear control. npj Systems Biology and Applications 8, 36 (2022)
JC Rozum. Attractor identification and control of Boolean and ODE network models in Systems Biology. PhD Dissertation in Physics, Pennsylvania State University (2022)
JC Rozum, D Deritei, KH Park, JGT Zañudo, R Albert. pystablemotifs: Python library for attractor identification and control in Boolean networks. Bioinformatics 38 (5), btab825 (2021)
JC Rozum, JGT Zañudo, X Gan, D Deritei and R Albert. Parity and time-reversal elucidate both decision-making in empirical models and attractor scaling in critical Boolean networks. Science Advances 7 (29), eabf8124 (2021)
JC Rozum and R Albert. Controlling the cell cycle restriction switch across the information gradient. Advances in Complex Systems 22 (07n08), 1950020 (2020)
D Deritei, JC Rozum, E Ravasz Regan, R Albert. A feedback loop of conditionally stable circuits drives the cell cycle from checkpoint to checkpoint. Scientific Reports 9 16430 (2019)
JC Rozum and R Albert. Identifying (un) controllable dynamical behavior in complex networks. PLOS Computational Biology 14 (12) (2018)
JC Rozum and R Albert. Self-sustaining positive feedback loops in discrete and continuous systems. Journal of Theoretical Biology 459 35-44 (2018)
JC Rozum. Classification of Five-dimensional Lie Algebras with One-dimensional Subalgebras Acting as Subalgebras of the Lorentz Algebra. Master’s Thesis in Mathematics, Utah State University (2015)
J Price, JC Rozum, G Ware, and D Baker. Global Nightly OH and O2 Mesospheric Airglow: Examining a Decade of Measurements Using the NASA SABER Satellite Sensor. Journal of the Utah Academy of Sciences, Arts, and Letters, 91 (2014)
Selected Talks and Poster Presentations
KH Park, FX Costa, LM Rocha, R Albert, JC Rozum. Robustness of biomolecular networks suggests functional modules far from the edge of chaos. Talk given at the International Conference on Systems Biology. Hartford CT, Oct. 10, 2023
F Xavier Costa, JC Rozum, AM Marcus and LM Rocha. Effective connectivity and bias entropy improve prediction of dynamical regime in automata networks. Talk given at the NetSci international conference. Vienna, Austria, Jul. 14, 2023
JC Rozum, JGT Zañudo, X Gan, D Deritei and R Albert. Parity and time-reversal in Boolean networks. Poster presented at the Cold Spring Harbor Laboratory Cellular Dyanamics and Models conference. Laurel Hollow, NY, May 19-21, 2021
JC Rozum. Boolean Networks: discrete dynamical models of high-dimensional nonlinear systems. Invited talk given at the University of Pennsylvania Mathematical Biology Seminar. Philadelphia, PA, Apr. 6, 2021
JC Rozum, JGT Zañudo, X Gan, and R Albert. Parity and time-reversal in Boolean networks. Talk given at the NetSci international conference. Rome, Italy, Sep. 25, 2020
JC Rozum and R Albert. Control robust trap spaces with applications to biomolecular networks. Talk given at the NetSci international conference. Burlington, VT, May 29, 2019
JC Rozum and R Albert. Identifying (un)controllable dynamical behavior with applications to biomolecular networks. Invited talk given at Pennsylvania State University Theoretical biology seminar. State College, PA, Feb. 20, 2018